Unveiling Biological Complexity: A Comprehensive Guide to UMAP in RNA Sequencing
Related Articles: Unveiling Biological Complexity: A Comprehensive Guide to UMAP in RNA Sequencing
Introduction
With great pleasure, we will explore the intriguing topic related to Unveiling Biological Complexity: A Comprehensive Guide to UMAP in RNA Sequencing. Let’s weave interesting information and offer fresh perspectives to the readers.
Table of Content
Unveiling Biological Complexity: A Comprehensive Guide to UMAP in RNA Sequencing

RNA sequencing (RNA-Seq) has revolutionized biological research by providing a detailed snapshot of gene expression within a cell or tissue. This technology allows scientists to quantify the abundance of RNA transcripts, revealing the intricate interplay of genes in various biological processes. However, the sheer volume of data generated by RNA-Seq poses a significant challenge in data analysis and visualization.
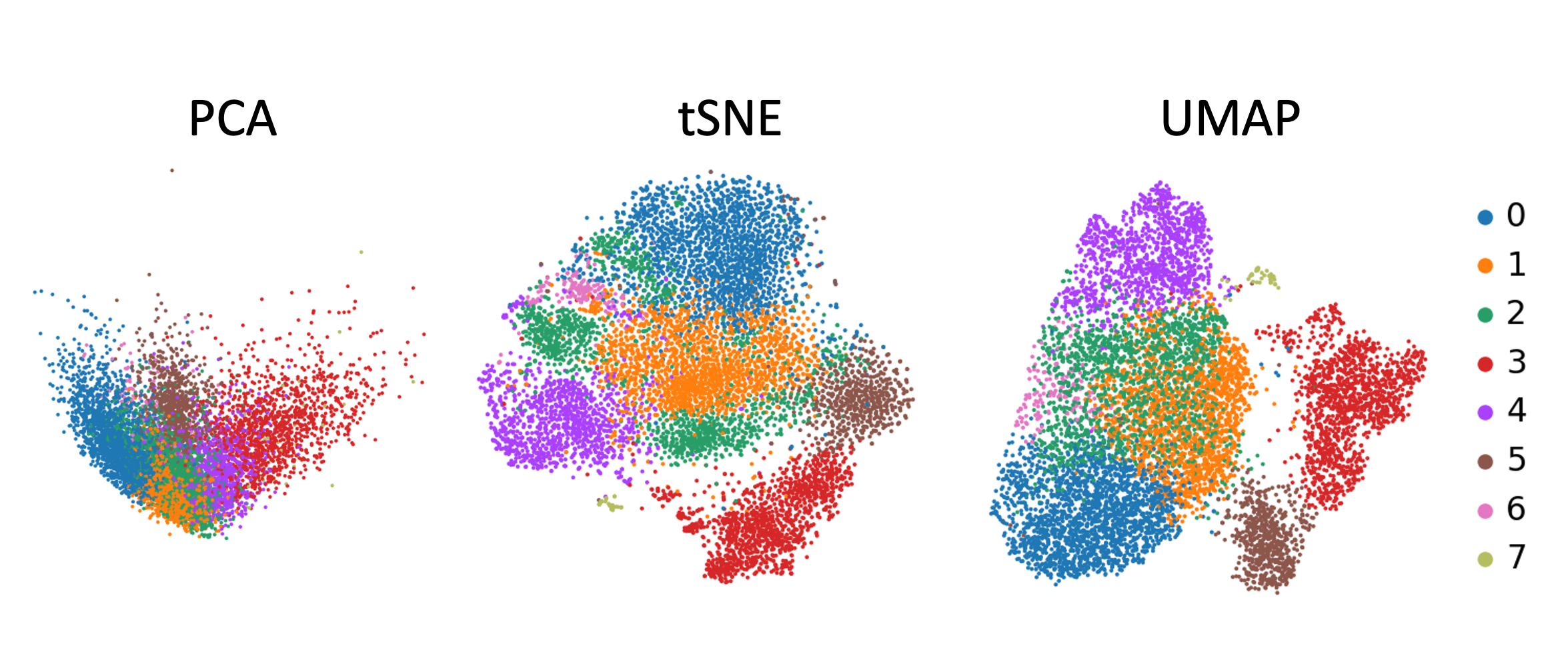
This is where Uniform Manifold Approximation and Projection (UMAP) emerges as a powerful tool, enabling researchers to navigate the complex landscape of RNA-Seq data and extract meaningful biological insights. UMAP is a dimensionality reduction technique that excels at preserving the intricate relationships between data points, transforming high-dimensional data into a lower-dimensional representation while retaining its essential structure.
Understanding the Power of UMAP in RNA-Seq
Imagine trying to decipher a complex tapestry woven with countless threads. Each thread represents a gene, and the intricate patterns formed by their interwoven relationships reveal the underlying biological processes. However, the sheer number of threads makes it impossible to discern the overall design. This is where UMAP comes into play.
UMAP acts like a skilled weaver, carefully disentangling the threads to reveal the underlying patterns. It projects the high-dimensional RNA-Seq data onto a lower-dimensional space, typically 2 or 3 dimensions, while preserving the crucial relationships between genes. This allows researchers to visualize the complex gene expression landscape and identify distinct clusters of cells or tissues based on their unique gene expression profiles.
Benefits of UMAP in RNA-Seq Analysis
The application of UMAP in RNA-Seq analysis offers several significant benefits:
- Enhanced Visualization: UMAP transforms the high-dimensional data into a visually comprehensible format, allowing researchers to easily identify patterns, clusters, and outliers within the gene expression landscape. This facilitates a deeper understanding of the underlying biological processes and reveals hidden relationships between genes.
- Improved Clustering Accuracy: UMAP excels at preserving the local neighborhood structure of the data, leading to more accurate and biologically relevant clusters of cells or tissues based on their gene expression profiles. This allows researchers to identify distinct cell types, developmental stages, or disease states within a heterogeneous population.
- Identification of Novel Biological Insights: By revealing hidden patterns and relationships in the data, UMAP can help researchers uncover novel biological insights that might otherwise remain obscured. This could lead to the discovery of new biomarkers, pathways, or therapeutic targets.
- Enhanced Data Exploration: UMAP provides a powerful tool for exploring the intricate landscape of RNA-Seq data, enabling researchers to identify potential areas of interest and focus their further investigations. This facilitates a more efficient and insightful analysis process.
- Reduced Computational Burden: By reducing the dimensionality of the data, UMAP significantly reduces the computational burden associated with analyzing large RNA-Seq datasets. This allows researchers to process and analyze data more efficiently and effectively.
Applications of UMAP in RNA-Seq
UMAP has found widespread application in various fields of biological research, including:
- Cell Type Identification: UMAP enables researchers to identify distinct cell types within a heterogeneous population based on their unique gene expression profiles. This is particularly valuable in studies of complex tissues, such as the brain or immune system.
- Disease Diagnosis and Prognosis: By analyzing gene expression patterns in diseased tissues, UMAP can help identify potential biomarkers for disease diagnosis and prognosis. This can lead to earlier detection and more effective treatment strategies.
- Drug Discovery and Development: UMAP can be used to identify potential drug targets by analyzing the gene expression profiles of cells or tissues affected by a disease. This can accelerate the drug discovery process and lead to the development of more effective therapies.
- Developmental Biology: UMAP allows researchers to track changes in gene expression during development, providing insights into the complex processes that govern cellular differentiation and tissue formation.
- Evolutionary Biology: UMAP can be used to compare gene expression patterns across different species, revealing evolutionary relationships and identifying genes that are involved in specific adaptations.
FAQs about UMAP in RNA-Seq
1. How does UMAP work?
UMAP is a non-linear dimensionality reduction technique that aims to preserve the local neighborhood structure of the data. It works by constructing a graph representation of the data, where each node represents a data point and edges represent the relationships between points. This graph is then embedded into a lower-dimensional space, while preserving the topological structure of the original data.
2. What are the advantages of using UMAP over other dimensionality reduction techniques?
UMAP offers several advantages over other dimensionality reduction techniques, such as t-SNE or PCA. It excels at preserving the local neighborhood structure of the data, leading to more accurate and biologically relevant clusters. Additionally, UMAP is generally faster and more scalable than t-SNE, making it suitable for analyzing large datasets.
3. How do I implement UMAP in my RNA-Seq analysis?
UMAP can be implemented in various software packages, including R, Python, and MATLAB. Several libraries and packages are available, such as umap and umap-learn, which provide functions for applying UMAP to RNA-Seq data.
4. What are some common challenges associated with using UMAP in RNA-Seq analysis?
While UMAP offers significant benefits, it is important to be aware of potential challenges. These include:
- Parameter Tuning: UMAP requires careful parameter tuning to achieve optimal results. This involves selecting appropriate values for parameters such as the number of neighbors, the minimum distance, and the metric used to calculate distances between data points.
- Data Preprocessing: Proper data preprocessing is crucial for obtaining meaningful results from UMAP. This involves removing noise, scaling data, and selecting relevant features.
- Interpretation: Interpreting the results of UMAP can be challenging, particularly when dealing with complex datasets. It is important to consider the biological context and validate the results using other methods.
Tips for Effective UMAP Implementation in RNA-Seq
- Choose Appropriate Parameters: Experiment with different parameter values to find the optimal settings for your specific dataset.
- Perform Data Preprocessing: Remove noise and outliers, scale data, and select relevant features to improve the performance of UMAP.
- Validate Results: Validate the results of UMAP using other methods, such as clustering algorithms or biological validation experiments.
- Consider Biological Context: Interpret the results of UMAP in the context of the underlying biology.
- Visualize and Explore: Utilize visualization tools to explore the low-dimensional representation of the data and identify potential areas of interest.
Conclusion
UMAP has emerged as a transformative tool in RNA-Seq analysis, enabling researchers to navigate the complex landscape of gene expression data and extract valuable biological insights. By preserving the intricate relationships between genes, UMAP provides a powerful means for visualizing, clustering, and interpreting RNA-Seq data, leading to a deeper understanding of biological processes and the discovery of novel insights. As RNA-Seq technology continues to advance, UMAP will undoubtedly play an increasingly important role in unlocking the secrets of the genome and driving groundbreaking discoveries in various fields of biological research.






Closure
Thus, we hope this article has provided valuable insights into Unveiling Biological Complexity: A Comprehensive Guide to UMAP in RNA Sequencing. We hope you find this article informative and beneficial. See you in our next article!
